Development of Novel Computational Tools¶
The broad range of models developed and used in the group include approaches from applied mathematics, machine learning, cheminformatics, Bayesian statistics, and quantum chemistry.
We utilize a number of computer languages and software tools in our research to develop specialized tools, conduct simulations, carry out data processing, and perform visualizations. These languages and applications include python, R, MATLAB, NetLogo, MCSim, TensorFlow, scikit-learn, Smoldyn, CompuCell3D, MCell and CellBlender, Gaussian, MOPAC, GROMACS, RDKit, Monolix Suite.
Some of the specialized tools we have developed are described below.
kc-hits¶
The computational package kc-hits 1 productivity-enhancing tool for the diverse scientists and agencies involved in classifying chemicals for their carcinogenic potential. |
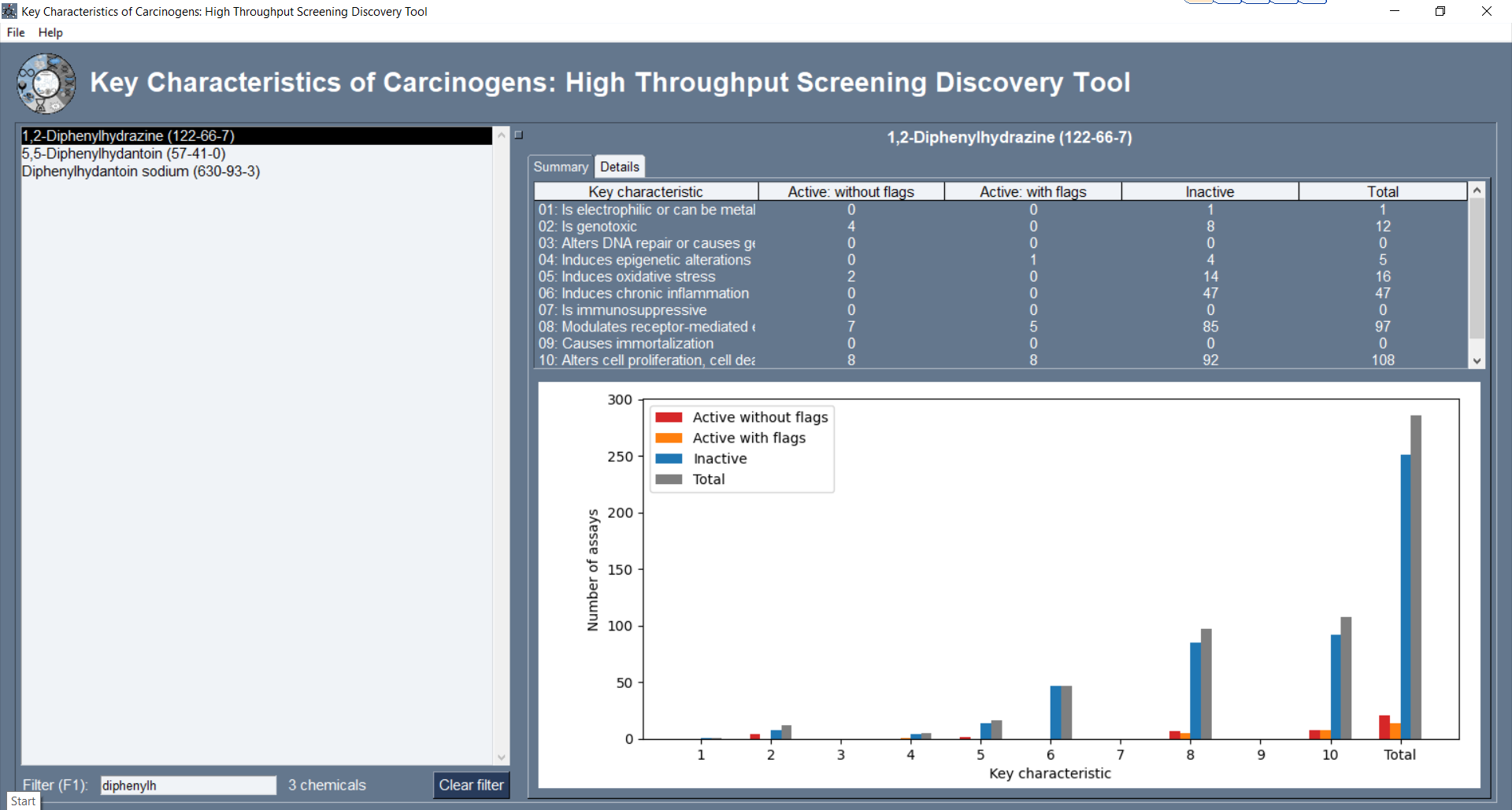
|
DoseSim¶
The computational package DoseSim 2 is useful in characterizing biomarkers and conducting dose estimation for various chemicals, including the organophosphorus insecticide, chlorpyrifos. |
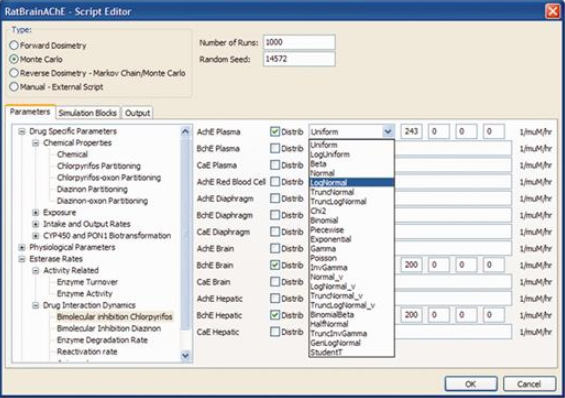
|
BioTRaNS¶
BioTRaNS 3 (Biochemical Tool for Reaction Network Simulation) is a package for a software package for computational xenobiotic metabolomics It has the capability to predict the metabolite inventory and pathways resulting from exposure to chemicals and mixtures of chemicals. |
|